SureSeq Myeloid MRD NGS Panel
Measurable residual disease of myeloid samples down to 0.05% VAF
- Expert-led, evidence-based and future-proof content
- Unparalleled uniformity and depth of coverage for difficult to sequence genes, e.g. CEBPA, NPM1 and FLT3-ITDs
- Rapid, streamlined workflow with Universal NGS Complete Workflow Solution
- Complimentary data analysis software with Interpret NGS Analysis Software
The SureSeqTM Myeloid MRD panel delivers
The SureSeqTM Myeloid MRD Panel has been designed in collaboration with leading cancer experts and in accordance with the European LeukemiaNet (ELN) recommendations [1,2] to offer a single, cost-effective NGS assay to investigate MRD in AML samples. The panel content incorporates key genes for assessing AML, including genes implicated in MPN and MDS, to investigate secondary AML progression.
In addition, genes related to research on potential drug response are included for truly comprehensive and informative sample analysis.
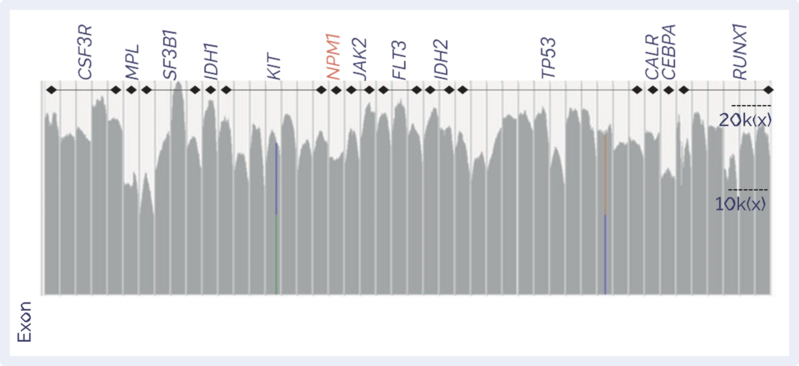
Utilising SureSeq’s intelligent panel design capabilities, the Myeloid MRD Panel surpasses the typical sensitivities offered by alternative NGS panels to accurately detect SNVs, indels and internal tandem duplications (ITDs) down to 0.05% VAF.
Gene targets
Gene | Exons |
CSF3R | Exons 13–17 |
MPL | Exon 10 |
| SF3B1 | Exons 13–16 |
IDH1 | Exon 4 |
| KIT | Exons 2, 8–11, 13 and 17 |
NPM1 | Exon 11 |
| JAK2 | Exons 12 and 14 |
| FLT3 | Exons 13-15 et 20 |
| IDH2 | Exons 4 and 5 |
| TP53 | Exons 2–11 (inc. NM_001276695:ex10, NM_001276696:ex10) |
| CALR | Exon 9 |
| Exon 9 | Exons 4–8 |
| CEBPA | Exon 1 |
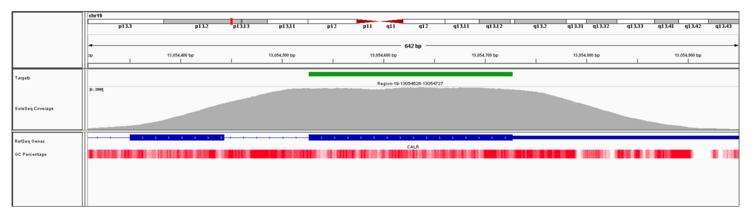
Figure 2: IGV plot showing high uniformity of coverage of all targeted regions in the panel, including NPM1 exon 11.
Figure 3: FLT3-ITDs of various sizes and even regions containing multiple ITDs can be confidently detected. ITD sizes are [A] 174 bp, [B] 225 bp, [C] 195 bp with additional 6 bp, [D] 120 bp, [E9 168 bp with additional 69 bp.
View all NGS library preparation kits & reagents
References
[1] European LeukaemiaNET. Available at: www.leukemia-net.org.
[2] Döhner et al Blood. 2022 ;140(12):1345-1377
Disclaimer
SureSeqTM: For Research Use Only, not for use in diagnostic procedures. Product availability may vary from country to country and is subject to varying regulatory requirements. Contact your local representatives for availability.
Manufacturer and Trademarks: SureSeqTM (Oxford Gene Technology IP Limited)
Technical information
| Feature | Specification | |
| Number of targets | 45 hotspot exons from 13 genes | |
| Panel size | 11.2 kb | |
| Mean target coverage | Up to 20,000x | |
| Limit of detection SNVs, indels, ITDs | 0.05% | 0.1% |
| DNA input recommended | 2 x 200 ng | 200 ng |
| Samples per run | ||
| NextSeq 500 High Output | 16 | 24 |
| NextSeq 2000 P3 | 48 | 72 |
| NovaSeq® SP | 32 | 48 |
| NovaSeq S1 | 64 | 96 |
Detection of SNVs, indels and an ITD, with expected frequency ranges of 0.1%-0.05%. SNVs are filtered to remove unique molecular identifiers (UMI) with a read family size of 1. A Myeloid Reference DNA Standard (Horizon Discovery) sequenced on a Illumina NextSeq® 500.
| Gene | Variant | Coordinate | 0.1% VAF | 0.1% VAF | 0.05% VAF | 0.05% VAF | Neg. ctrl | Neg. ctrl | |
| Read depth | Observed VAF | Read depth | Observed VAF | Read depth | Observed VAF | ||||
| SNV | SF3B1 | c.2219G>A | chr2:197401989 | 7426 | 0.13 | 14282 | 0.03 | 15076 | 0 |
| IDH1 | c.394C>T | chr2:208248389 | 6743 | 0.09 | 15572 | 0.06 | 15734 | 0 | |
| JAK2 | c.1849G>T | chr9:5073770 | 10408 | 0.12 | 22348 | 0.08 | 22065 | 0 | |
| FLT3 | c.2503G>T | chr13:28018505 | 9747 | 0.11 | 24098 | 0.04 | 23942 | 0 | |
| IDH2 | c.515G>A | chr15:90088606 | 7294 | 0.08 | 16554 | 0.04 | 18875 | 0 | |
| TP53 | c.722C>T | chr17:7674241 | 5166 | 0.12 | 12609 | 0.03 | 13769 | 0 | |
| Indel | NPM1 | c.860_863dup | chr5:171410539 | 8164 | 0.11 | 18630 | 0.07 | 17630 | 0 |
| JAK2 | c.1611_1616del | chr9:5070021 | 12270 | 0.03 | 28197 | 0.02 | 25411 | 0 | |
| ITD | FLT3 | ITD300 | chr13:28033909 | 6045 | ≥ 0.1 | 12265 | ≥ 0.05 | 10473 | 0 |
Ordering information
| Product | Contents | Cat. No. |
| SureSeq Myeloid MRD Complete NGS Workflow Solution V2 (48) | Enrichment baits sufficient for 12 hybridisation reactions. Bundle of 1 x Universal Library Preparation Kit (96) containing PCR primers and enzymes. 1 x Universal Hybridisation & Wash Kit V2 (96). 1 x Pre-PCR Universal Bead Kit (96). 1 x Post-PCR Universal Bead Kit (96). 1 x Universal Index Adapter Kit (96). Interpret NGS Analysis Software | 780126-48 |
| SureSeq Myeloid MRD Panel (48) | Enrichment baits sufficient for 12 hybridisation reactions. Interpret NGS Analysis Software | 770026-48 |
| Universal NGS Workflow Solution V2 (96) | Bundle of 1 x Universal Library Preparation Kit (96) containing PCR primers and enzymes, 1 x Universal Hybridisation & Wash Kit V2 (96). 1 x Pre-PCR Universal Bead Kit (96). 1 x Post-PCR Universal Bead Kit (96). 1 x Universal Index Adapter Kit (96) | 770510-96 |
Sysmex Europe SE
Bornbarch 1
22848 Norderstedt
Germany
+49 (40) 527 26 0
+49 (40) 527 26 100
Product documents
Regulatory documents
Regulatory documents, such as Instructions for Use, can be accessed with a valid My Sysmex login:
Go to My Sysmex